Abstract
Chronic lymphocytic leukemia (CLL) is characterized by constitutively activated B cell receptor (BCR) signaling. Inhibitors of BCR signaling such as ibrutinib and idelalisib have proved to be highly efficacious in CLL patients. While the molecular basis for resistance to ibrutinib has been elucidated, mechanisms leading to idelalisib resistance are unknown. We have previously generated resistance to PI3K-δ inhibitor in vivo (Scheffold et al., ASH 2016) and identified IGF1R to be uniformly upregulated in resistant tumors.
Here, we further characterized the contribution of IGF1R to PI3K-δ inhibitor resistance. Overexpression of IGF1R in A20 cells which are highly sensitive to iPI3K-δ resulted in a drastic reduction in response to iPI3K-δ (IC50 40µM vs. 126nM). IGF1R overexpression was associated with a strong increase in p-ERK levels indicating activation of MAPK signaling. Targeting IGF1R using the specific inhibitor linsitinib sensitized the IGF1R overexpressing A20 cells to iPI3K-δ, accompanied by a decrease in p-ERK levels. Additionally, long term treatment of MEC1 and MEC2 cells with increasing doses of idelalisib resulted in upregulation of IGF1R and MAPK signaling in RNA and protein levels. Furthermore, analysis of paired RNA samples from idelalisib resistant patients, collected before start of idelalisib treatment (N=6) and upon disease progression under therapy (N=9) showed elevated expression of IGF1R at baseline and upon disease progression compared to a reference sample set (N=27) of previously untreated CLL, highlighting a potential role of IGF1R signaling in iPI3K-δ resistance.
The presence of high IGF1R levels in the idelalisib resistant samples already at baseline before treatment led to the hypothesis that receptor tyrosine kinases (RTKs) could define patient subsets early in the disease course, impacting response to various treatments. Therefore, gene expression profiling was performed on RNA from 337 previously untreated CLL patient samples from the CLL8 trial of the GCLLSG, comparing fludarabine+cyclophosphamide (FC) vs. FC+Rituximab (FCR). Affymetrix Human Exon 1.0 ST Arrays was used for the analysis and data for core probe sets and expression levels of RTKs such as IGF1R, IGF2R, INSR, FGFR1, FGFR2, FGFR3, FGFR4, ROR1, ROR2, DDR1, DDR2, LTK, AXL, PDGFRA, PDGFRB, ERBB2, ERBB3, ALK, TIE1 and RET, all known to be expressed in hematopoietic cells were analyzed. Strikingly, unsupervised clustering identified 2 major clusters that differed mostly with respect to IGF1R expression.
To understand the clinical relevance of IGF1R in CLL8, associations with disease characteristics and survival were analyzed after dichotomization into IGF1Rhigh (>median) and IGF1Rlow (<median) expression subgroups. Among the clinical characteristics, IGF1R expression was not associated with age, ECOG status, presence of B-symptoms and CIRS score. However, high expression of IGF1R was significantly associated with male sex (P=0.002), Binet stage B (P=0.001), high serum thymidine kinase (P<0.001) and the high /very high CLL-IPI risk groups (P<0.001). Among the genetic characteristics, high IGF1R expression was significantly associated with unmutated IGHV (P<0.001). Interestingly, among the hierarchical genomic aberrations subgroups, high IGF1R expression was significantly associated with trisomy 12 (P=0.015). On the contrary, 13q- as a sole abnormality showed a trend towards association with low IGF1R expression (P=0.065).
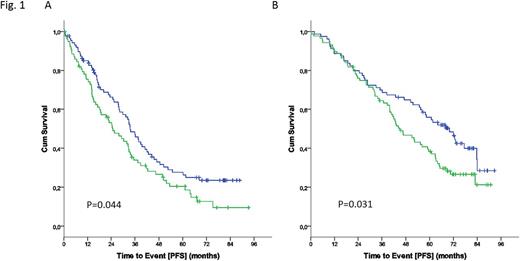
At a median observation time of 70.3 months, there were 234 events for progression free survival (PFS) and 120 events for overall survival (OS). Strikingly, high IGF1R expression was significantly associated with poor PFS in both the FC (Fig. 1A: HR 1.433; 95%CI 1.008-2.037; P=0.045) and FCR (Fig. 1B: HR 1.518; 95%CI 1.036-2.225; P=0.031). Though IGF1Rhigh is associated with poor PFS irrespective of the treatment arm, addition of rituximab to FC proved beneficial in both the IGF1Rlow (median: FC 33.4 vs. FCR 69.9 months; P=0.002) as well as IGF1Rhigh (median: FC 24.7 vs. FCR 44.9 months; P<0.001) subgroups.
In summary, IGF1R is a RTK of potential pathogenic and prognostic relevance that is upregulated in a subset of CLL cases and is associated with poor prognosis in the chemoimmunotherapy setting. Additionally, signaling through IGF1R may compensate for PI3K-δ inhibition, thereby contributing to drug resistance that can be overcome by specific targeting.
Bahlo:F. Hoffmann-LaRoche: Honoraria, Other: travel grants. Fischer:Roche: Other: Travel Grants. Hallek:Abbvie: Consultancy, Honoraria, Research Funding; Janssen: Consultancy, Honoraria; Roche: Consultancy, Honoraria, Research Funding, Speakers Bureau. Döhner:Boehringer Ingelheim: Research Funding; Celator: Honoraria; Novartis: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Seattle Genetics: Honoraria; Sunesis: Honoraria; Celgene: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Pfizer: Research Funding; Bristol Myers Squibb: Research Funding; Arog Pharmaceuticals: Honoraria, Research Funding; Astex Pharmaceuticals: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Amgen: Honoraria; Agios: Honoraria; Abbvie: Honoraria. Stilgenbauer:Novartis: Consultancy, Honoraria, Research Funding; Genentech: Consultancy, Honoraria, Research Funding; Amgen: Consultancy, Honoraria, Research Funding; Janssen: Consultancy, Honoraria, Research Funding; Mundipharma: Consultancy, Honoraria, Research Funding; Gilead: Consultancy, Honoraria, Research Funding; Genzyme: Consultancy, Honoraria, Research Funding; Sanofi: Consultancy, Honoraria, Research Funding; Boehringer-Ingelheim: Consultancy, Honoraria, Research Funding; GSK: Consultancy, Honoraria, Research Funding; Pharmacyclics: Consultancy, Honoraria, Research Funding; Celgene: Consultancy, Honoraria, Research Funding; AbbVie: Consultancy, Honoraria, Research Funding; Hoffman La-Roche: Consultancy, Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal